A third
toolbar now offers options for working with SIMS spectra. The buttons are
mostly for stepping and zooming into spectra where the background is typically
close to zero.

Figure 1 New SIMS toolbar shown below
the two previously existing toolbars.
When
viewing SIMS spectra, zoom operations typically define the maximum intensity
and the range over which the display should focus, whereas XPS/AES spectra
differ in that the peaks emerge from a background of counts and therefore
require zoom operations to allow for a minimum intensity as well as a maximum
to be specified. A toolbar button on the SIMS toolbar ![]() switches
between these two modes of zoom operations. The default for SIMS spectra
ensures that all zoom operations are relative to a zero base line; in this
mode, a line is drawn along the bottom of the Active Tile. XPS/AES spectra on the
other hand, when initially displayed are viewed relative to the minimum counts
determined from the data.
switches
between these two modes of zoom operations. The default for SIMS spectra
ensures that all zoom operations are relative to a zero base line; in this
mode, a line is drawn along the bottom of the Active Tile. XPS/AES spectra on the
other hand, when initially displayed are viewed relative to the minimum counts
determined from the data.
The display
option toggled by the zoom mode toolbar button is also available on the Y Axis
property page on the Tile Display Parameters dialog window (Figure 2). A
logarithmic display is also enabled from the same property page seen in Figure
2 and can be switched on/off using the SIMS toolbar button![]() .
.
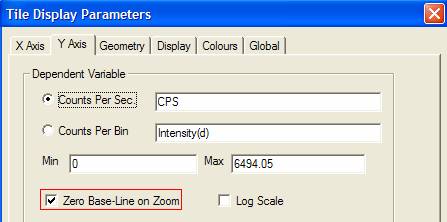
Figure 2: Tile Display Parameter
Dialog Window.
The SIMS
toolbar buttons provide a means of altering the interval displayed in the
Active Tile.
![]() The mass interval is cycled through a
predefined set of ranges. The mass interval relative to the current minimum
mass in the Active Tile is adjusted in order to display 5, 10, 25, 50 and 100 amu intervals.
The mass interval is cycled through a
predefined set of ranges. The mass interval relative to the current minimum
mass in the Active Tile is adjusted in order to display 5, 10, 25, 50 and 100 amu intervals.
![]() Step the mass range left or right by half the
current mass range in the Active Tile. The intensity range is not rescaled
unlike the equivalent buttons
Step the mass range left or right by half the
current mass range in the Active Tile. The intensity range is not rescaled
unlike the equivalent buttons ![]() found on the Options Toolbar initially above
the SIMS toolbar.
found on the Options Toolbar initially above
the SIMS toolbar.
![]() Expand the mass range without rescaling the
intensity interval. Again, the equivalent button on the Options Toolbar
rescales the intensity scale.
Expand the mass range without rescaling the
intensity interval. Again, the equivalent button on the Options Toolbar
rescales the intensity scale.
![]() Step left and right by one amu.
The intensity is rescaled with each step.
Step left and right by one amu.
The intensity is rescaled with each step.
![]() The spectrum displayed in the Active Tile
is divided, in terms of the mass scale, into a set of new tiles, the number of
which is specified on the dialog window invoked when the button is pressed.
These new tiles appear at the bottom of the current scrolled list of display
tiles and are therefore in a state ready for printing.
The spectrum displayed in the Active Tile
is divided, in terms of the mass scale, into a set of new tiles, the number of
which is specified on the dialog window invoked when the button is pressed.
These new tiles appear at the bottom of the current scrolled list of display
tiles and are therefore in a state ready for printing.
![]() The mass intervals defined by the current set
of quantification regions are copied into a new Experiment Frame. This is a
means of extracting smaller units of data for transfer to other programs or for
use with features such as peak fitting within CasaXPS.
The mass intervals defined by the current set
of quantification regions are copied into a new Experiment Frame. This is a
means of extracting smaller units of data for transfer to other programs or for
use with features such as peak fitting within CasaXPS.
![]() Calibrates the set of spectra displayed in the
Active Tile.
Calibrates the set of spectra displayed in the
Active Tile.
Time binned
spectra are calibrated with respect to mass by first defining a set of
quantification regions, where each region specifies the location of a peak and
the mass formula for the ion responsible for the peak position within the time
domain. The mass formula is entered into the region fields labelled Name and
the Start and End fields must be adjusted to include the peak maximum within
the region. If calibration regions are defined on more than one spectrum,
overlaying these spectra in the Active Tile before pressing the ![]() toolbar button will calibrate each of the
displayed spectra.
toolbar button will calibrate each of the
displayed spectra.
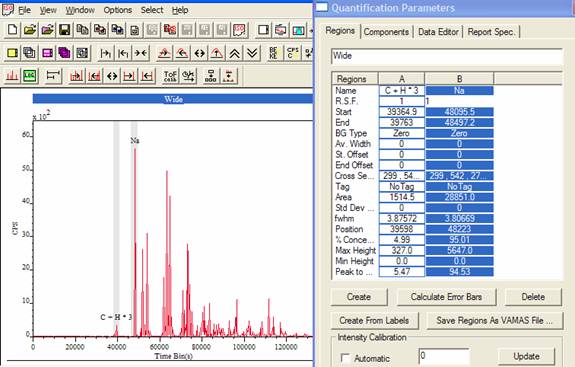
Figure 3 Time binned spectrum showing two peaks defined in preparation for mass calibration.
In order to calibrate a time-binned spectrum, two or more peaks must be defined before the toolbar button is pressed. A quadratic relationship between the positions of the peaks in the time domain and the same peaks in the mass domain is established from the information provided by the quantification regions. After calibration the spectrum is displayed with a mass scale, however the peaks remain in the time domain while the abscissa is labeled with respect to the mass domain. The element library will place markers on the spectrum in the correct mass position once the spectrum is calibrated, however prior to calibration the element markers are not valid.
Once calibrated, time binned spectra may be converted into
mass binned spectra using the ![]() toolbar button. On pressing the toolbar button
a dialog window appears, where the number of mass bins used in the conversion
must be specified.
toolbar button. On pressing the toolbar button
a dialog window appears, where the number of mass bins used in the conversion
must be specified.
Converting a time-binned spectrum to a mass-binned spectrum involves a transformation that causes information to be lost towards lower masses and where there is missing information at higher masses, depending on the number of mass bins chosen. This is a consequence of the non-linear relationship between the time and mass position of peaks. Three different strategies are used to perform this transformation and so after the operation is performed three spectra appear in the mass-binned equivalent of the time-binned spectrum (offered in a new Experiment Frame):
1) The counts within a time bin are allocated to whichever mass bin the nominal time maps to in the mass domain, then the average of these counts is entered into the new mass-bin. The spectrum is then scaled in an attempt to account for the change-of-variable required for integration of the charge under a peak when switching between the time and mass domains.
2) Each mass-bin is the sum of all counts taken from time-bins falling into the given mass-bin under the current time-to-mass calibration. No scaling is performed, nor any smoothing to account for the number of time-bins falling within the mass-bin varying as a function of mass or number of mass-bins selected.
3) Conversion is performed assuming the time-binned spectrum can be used to interpolate the intensity at each discrete time associated with the mass-bins. The spectra are scaled according to the same regime as 1) to account for the change-of-variable within peak integration calculations. The success of this method is dependent on the choice for the number of mass-bins used in the conversion.
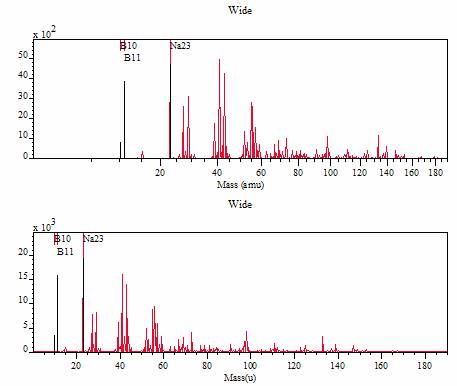
Figure 4 Spectrum after calibration (top) and following time-to-mass re-binning (bottom). Note the mass scale is linear after re-binning but non-linear in the time-binned spectrum. Note also that the relative intensity of peaks alters after re-binning the counts.
A default mass library offers a set of isotopic mass values for identifying elemental mass peaks, while the Exact Mass Calculator property page on the Element Library dialog window offers a means of computing masses for molecular ions. Once a set of lines is constructed using the Exact Mass Calculator, the mass values may be saved to file or transferred into the current element library.
Figure 5 shows molecular mass formulae prepared using the Exact Mass Calculator, where peaks associated with the C2Hn sequence are identified. Arithmetic expressions involving the elemental masses are used to define the desired molecular mass; markers appear on the spectrum at the computed mass value, which can be toggled on and off in a similar way to the Element Table scrolled list. Once a set of lines such as those seen in Figure 5 are prepared, these lines can be saved to file and recalled later using the Save and Load buttons on the Exact Mass Calculator property page. In addition, to saving and restoring molecular tables of masses, sets of masses can be transferred into the current element library and associated with a given element (Move to Lib button on the Exact Mass Calculator property page). The result of selecting an element from the periodic table is that the molecular peak markers will be displayed whenever the chosen element is selected on the periodic table. For example, the molecular peaks shown in Figure 5 could be associated with carbon and so these peak-markers would appear whenever carbon is selected from the periodic table or from the Element Table property page.
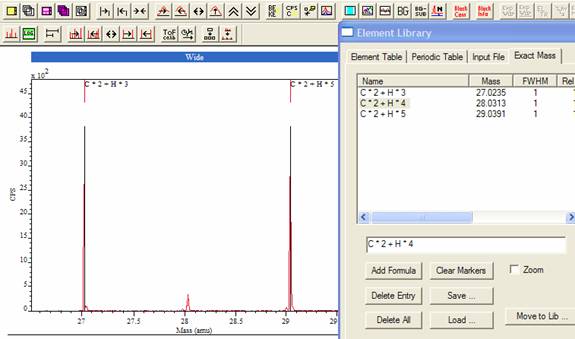
Figure 5: Exact Mass Calculator
To add a formula to the Exact Mass Calculator table, enter the expression into the text-field below the table (Figure 5) and then press the Add Formula button. Entries in the table can be removed by selecting the unwanted entry in the table, followed by pressing the Delete Entry button. All entries are removed when the Delete All button is pressed; this action is in contrast to the Clear Markers button, which removes all the peak markers from the display rather than the table. If the formulae are selected within the Exact Mass Table, then the molecular markers will again appear on the spectrum or will be removed from the display if already present.
A further action associated with molecular marker selection in the scrolled table occurs when the Zoom tick-box is checked. When checked, the result of selecting a molecular formula in the table is the display mass range is adjusted to span the selected mass position.
A new form of annotated text can appear on spectra, where a chemical formula is interpreted as subscripted and superscripted text (Figure 6). The arithmetic symbols are removed from the formula, where multiplication causes the following text to appear as a subscript, while text inside “<” and “>” symbols are displayed as superscripts. A tick box labeled Formula (Figure 6) enables this display option. Figure 6 shows two pairs of text, with and without the Formula tick box active when the Apply button is pressed on the Text property page shown.
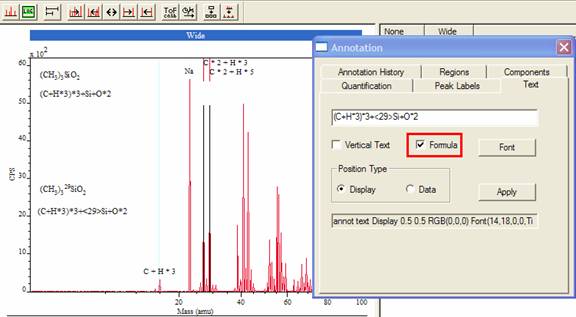
Figure 6