Marking Pixels and Spectra
Summation
Visualising
the surface of a sample in terms of chemical state images is a goal in itself;
however the ability to construct spectra from the individual spectra-at-pixels
corresponding to a range of pixel intensities represents a means of examining
the related chemistry in detail. Further, while PCA provides a means of
examining the spectra at each pixel, the procedure does require the judicious
choice of the information baring abstract factors and ultimately a least square
criterion fits the abstract factors to the raw spectra-at-pixels; as such
errors are possible, therefore a means of summing spectra from similar pixels
provides both a useful tool and also an independent approach for generating
spectra from the raw data in image data sets with an acceptable signal-to-noise
for peak-fitting and quantification.
The
procedure in CasaXPS for summing spectra-at-pixels is based on the definition
of a false-colour visualisation of an image, where various intensity ranges are
assigned different pixel colours. The resulting colour scale allows the pixels
within an image to be partitioned into zones of identical chemical state
information identified by a pixel colour and using the false-colour image, the
raw spectra-at-pixels are summed to produce as many spectra from an image data
set as there are false colours in the display of the image. The steps en route
to summing the spectra-at-pixels are as follows:
- Acquire an image data set in
which images are acquired at regular energy interval spanning all the
photoelectric lines required for quantifying the sample surface.
- Use PCA to create a set of
images representative of the chemical state information from the sample.
- Define a false-colour scale for
an image such that the surface is identified using one or more intensity
ranges and colours.
- Define the false-colour scale
as the active template for summing the pixel spectra.
- For each spectra-at-pixels
Experiment Frames generated from the original image data sets, apply the
false-colour scale template. The result of applying the template to an
Experiment Frame containing spectra-at-pixels is a new Experiment Frame
containing the spectra representing the sum of all the pixel spectra
corresponding to each of the false-colours in the template image.
The details
for performing these tasks now follow, where the steps are illustrated using
the example of an MRS-3 standard and data take from a Kratos Axis Ultra.
Step 1: Images
were acquired at energies spanning the Cr 2p doublet, O 1s, C 1s, Sn 3d and In 3d photoelectric lines (Figure 1).
Step 2: For this example, the Cr 2p images are
converted to spectra at each pixel (Figure 2). At a pixel level, the
signal-to-noise is poor, however by using PCA to partition the signal from the
noise throughout the entire set of spectra (256x256 spectra), the data at each
pixel is enhanced to permit the intensity at each pixel for the Cr 2p regions
to be defined using a quantification region. The details of the PCA analysis is
discussed elsewhere in the manual (see image processing section). However, the
result of the PCA is shown in Figure 3.
Step 3: The image in Figure 3 in visualised using
three false-colours. The intensity ranges, as seen via the colour scale in
Figure 3, are defined using the Colour
Scale property page on the Image Processing dialog window. The cursor is
used to drag out a line on over the image from which the intensities at the end
points of the line are available for use on the dialog window invoked by the Add False Colour button. Alternatively,
dragging out a box over the colour scale displayed next to the image will also
define a range of intensities, the limits of which will appear in the
text-fields on the resulting dialog window. The intensities of the pixels at
the end points of the line or from the box over the colour scale are entered on
the Image Lookup Table Range dialog
window, which appears when the Add False
Colour button is pressed; these values can be manually altered using the
edit text-fields. The colour used to represent the intensity range can also be
chosen from the Image Lookup Table Range
dialog window, however if the default colour is used rather than choosing a
new one, the colour scale colours and the colour used to plot spectra, will be
identical (assuming the default order of selection before the spectra are
overlaid in the Active Display tile).
On pressing
the Add False Colour button, a
processing command is added to the processing history. Adjustments to the
processing history entries can be made in the usual fashion using the
Processing History property page on the Spectrum Processing dialog window.
Step 4: Once the false-colour scale is prepared as
seen in Figure 3, the next step is to define the image as the template for
generating spectra. The Image Processing property page on the Image Processing
dialog window provides the means of specifying the image plus false-colour
scale for use with the spectra-at-pixels created from the image data set.
Display the false-colour image in the Active Tile and press the button labelled
Define Image. The image will redraw
in the Active Tile, which indicates that the definition has been achieved.
Step 5: Following the definition of the template
image, any spectra-at-pixel files containing only those VAMAS blocks from the original
creation step can be manipulated using the template image. Switch focus to the
Experiment Frame containing the spectra-at-pixels and press the button labelled
Sum Spectra Using Colours, also on
the Image Processing property page. A new Experiment Frame is created in which
a new VAMAS block (one for each colour used in the false-colour scale)
containing the sum of all the spectra-at-pixels with identical colour
assignments (Figure 4).
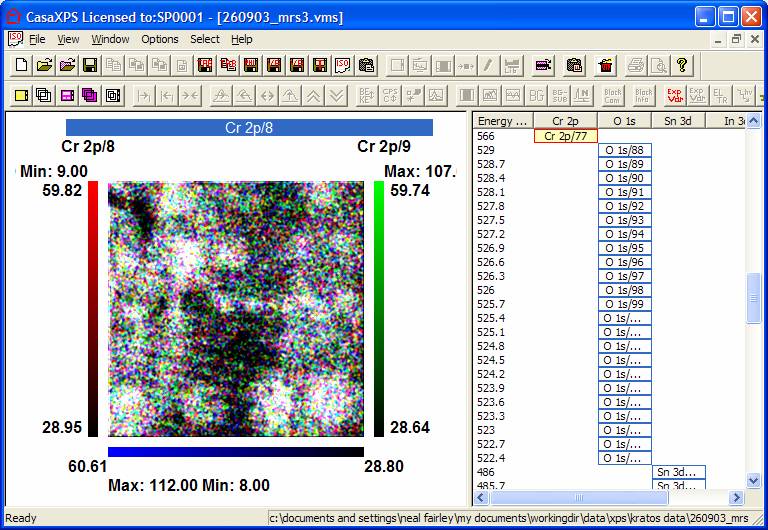
Figure 1: Image data set from which spectra at pixels are generated.
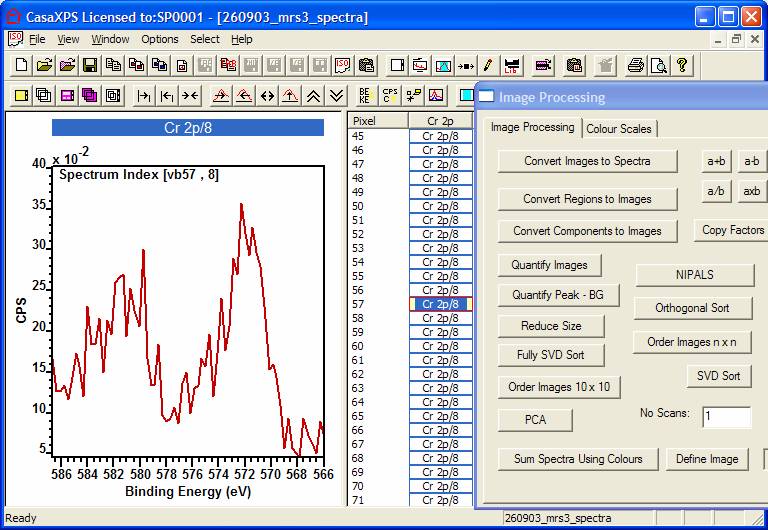
Figure 2: An example of a spectrum from a pixel
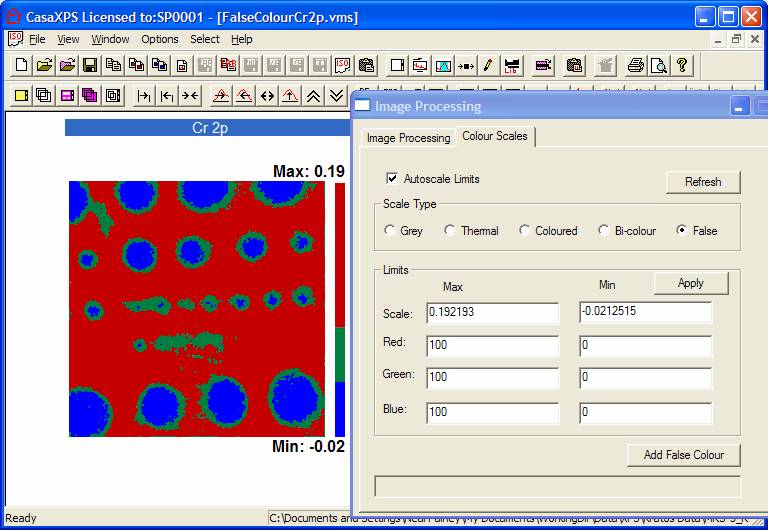
Figure 3: False Colour Scale
highlighting the regions of interest.
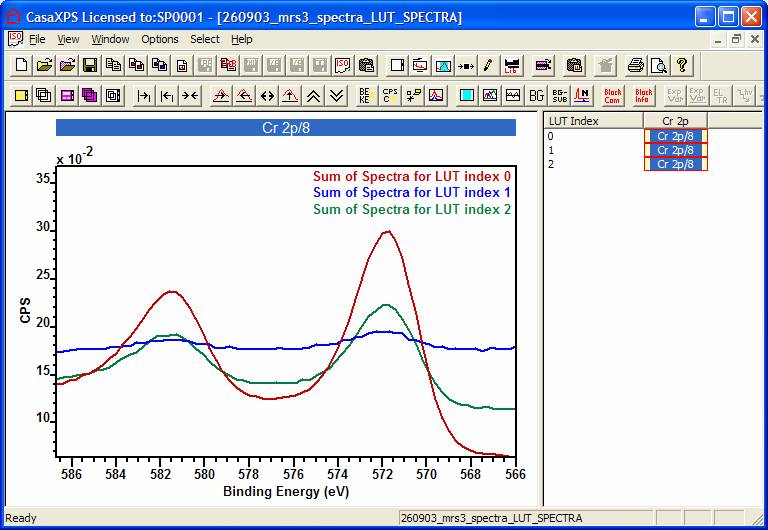
Figure 4: Spectra summed from spectra
at pixels for each of the three colour zones shown in Figure 3.